† co-first authors. *(co-)corrisponding authors. PL and trainees from the lab are in bold.
-
Dindo, M.; Metson, J.; Ren, W.; Chatzittofi, M.; Yagi, K.; Sugita, Y.*; Golestanian, R.*; Laurino, P.* Enzymes can activate and mobilize the cytoplasmic environment across scales 2025 Submitted https://www.biorxiv.org/content/10.1101/2025.01.28.635259v1
-
Bevilacqua, A.; Rios Maciel, R.; Pascarelli, S.; Dindo, M.; Shen, Q. A.*; Laurino, P. * Enzyme Activity Regulates Substrate Diffusion by Modulating Viscosity in Crowded Milieu. 2024 Submitted https://www.biorxiv.org/content/10.1101/2024.09.28.615560v1
-
Kotsifaki, D. G.∗; Truong, V. G.; Dindo, M.; Laurino, P.; Nic Chormaic, S.∗ Hybrid Metamaterial Optical Tweezers for Dielectric Particles and Biomolecules Discrimination. Submitted. arXiv 2024 https://arxiv.org/abs/2402.12878
-
Muthahari, A. Y.; Aditama, R.; Providaki, M.; Tsirigotaki, A.; Sarafoglou, C.; Xu, R.; Krishnamurthy, S.; Hertadi, R.; Kokkinidis, M.; Hui, C.; Laurino, P.; Pozidis, C.*; Gouridis, G* Thermodynamic principles govern evolutionary tradeoffs by regulating allostery. Submitted 2023
2025
26. Dindo, M.*; Conter, C.; Uechi, G.; Pampalone, G.; Ruta, L.; Pey, A. L.; Rossi, L.; Laurino, P.; Magnani, M.; Cellini, B.* Engineered Oxalate decarboxylase boosts activity and stability for biological applications. ACS Omega 2025 just accepted https://www.biorxiv.org/content/10.1101/2024.05.06.592502v1
25. Demkiv, A. O., Toledo-Patiño, S.; Medina Carmona, E.; Berg, A.; Pinto, G. P.; Parracino, A.; Sanchez-Ruiz, J. M.; Hengge, A. C.; Laurino, P.; Longo, L. M. * and Kamerlin, S. C. L.* Redefining the Limits of Functional Continuity in the Early Evolution of P-Loop NTPases. Molecular Biology and Evolution 2025 https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msaf055/8071345 just accepted
24. Muthahari, Y. A.†; Magnus, L.†; Laurino, P.*, From Duplication to Fusion: Expanding Dayhoff’s Model of Protein Evolution. Protein Science 2025 https://onlinelibrary.wiley.com/doi/10.1002/pro.70054 Perspective just accepted
2024
23. Matthews, M. M., Kim, T. G.; Kim, K. Y.; Meshcheryakov, V.; Iha, H. Y.; Tama, M.; Sasaki, D.; Laurino, P.; Toledo-Patiño, S.; Collins, M.; Hsieh, T.-Y.; Shibata, S.; Shibata, N.; Obata, F.; Fujii J.;, Ito, F.; Ito, H.; Ishikawa, H.; Wolf, M.* Engineered protein subunit COVID19 vaccine is as immunogenic as nanoparticles in 2 mouse and hamster models. Scientific Reports 2024, 14, 25528.
22. Ochiai, Y.; Clifton, B. E.; Le Coz, M.; Terenzio, M.; Laurino, P.* SUPREM: an engineered non-site-specific m6A RNA methyltransferase with highly improved efficiency. Nucleic Acids Research 2024 52, 20,12158–12172t https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae887/7824607
21. Clifton, B. E.*; Alcolombri, U.; Uechi, G-I; Jackson, C.; Laurino, P.* The ultra-high affinity transport proteins of ubiquitous marine bacteria. Nature 2024, 634, 721- 728 https://www.nature.com/articles/s41586-024-07924-w
20. Weronika Jasinska†, Mirco Dindo†, Sandra M. Correa, Adrian W.R. Serohijos, Paola Laurino*, Yariv Brotman*, Shimon Bershtein* Non-consecutive enzyme interactions within TCA cycle supramolecular assembly regulate carbon-nitrogen metabolism. Nature Communications 2024 15, 5285 https://www.nature.com/articles/s41467-024-49646-7
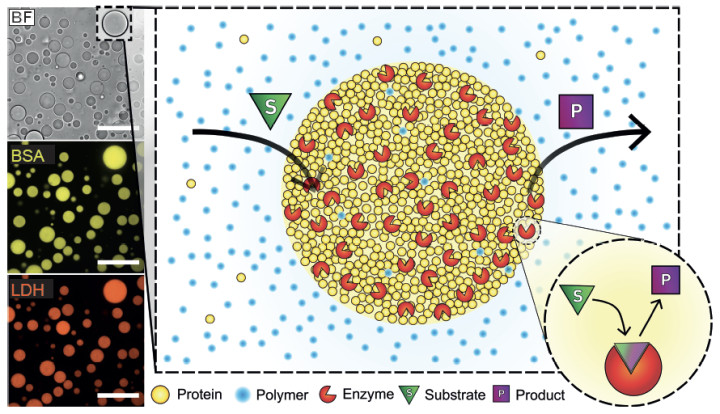
19. Dindo, M.†; Bevilacqua, A.†; Soligo, G.; Calabrese, V.; Monti, A.; Shen, Q. A.; Rosti, M. Laurino, P.* Chemotatic interactions drive migration of membraneless active droplets. Journal of American Chemical Society 2024 146, 23, 15965–15976 https://pubs.acs.org/doi/10.1021/jacs.4c02823
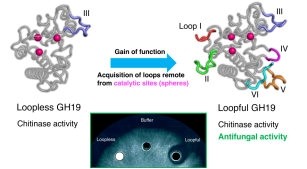
18. Kozome, D.; Sljoka, A.; Laurino, P.* Remote loop evolution reveals a complex biological function for chitinase enzymes beyond the active site. Nature Communications 2024, 15, 3227 https://rdcu.be/dELQk
2023
17. Gade, M.; Gardner, J.; Jain, P.; Laurino, P.* Nucleotide-driven specifisity of DNA Methyltransfersase ChemBioChem, 2023 24, e2023000 https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cbic.202300094
Paola was interviewed in Nature Catalysis to discuss the Roz Inclusivity project: "Helping Hands"
2022
16. Dindo, M.; Bevilacqua, A.; Laurino, P.* Enzymes and liquid-liquid phase separation: a new era for the regulation of enzymatic activity. Review, 2022, SEIBUTSU BUTSURI Vol. 63 No. 1(通巻365号)https://www.biophys.jp/journal/journal_dl.php?fnm=63-1
15. Toledo-Patiño, S.; Pascarelli, S.; Uechi, G.; Laurino, P.* Insertions and deletions mediated functional divergence of Rossmann fold enzymes. PNAS, 2022, 119, 48, e2207965119 https://www.pnas.org/doi/10.1073/pnas.2207965119
14. Jayaraman, V. †; Toledo-Patiño, S.†; Noda-García, L.*; Laurino, P.* Mechanisms of Protein evolution Protein Science, 2022, 31, e4362. https://onlinelibrary.wiley.com/doi/10.1002/pro.4362
13. Dindo, M.; Pascarelli, S.; Chiasserini, D.; Grotelli, S.; Constantini, C.; Uechi, G.; Giardina, G.*; Laurino, P.*; Cellini B.* Structural dynamics shape the fitness window of alanine:glyoxylate aminotransferase. Protein Science, 2022, 31, e4303. https://onlinelibrary.wiley.com/doi/10.1002/pro.4303
12. Pascarelli, S.; Laurino, P.* Inter-paralog aminoacid inversion events in large phylogenies of duplicated proteins Plos Comp Biol 2022, 18, 4, e1010016. https://doi.org/10.1371/journal.pcbi.1010016
11. Clifton, B. E.; Kozome, D.; Laurino. P.* Efficient exploration of sequence space by sequence-guided protein engineering and design ACS Biochemistry, 2023 62, 2, 210-220 https://pubs.acs.org/doi/10.1021/acs.biochem.1c00757
2021
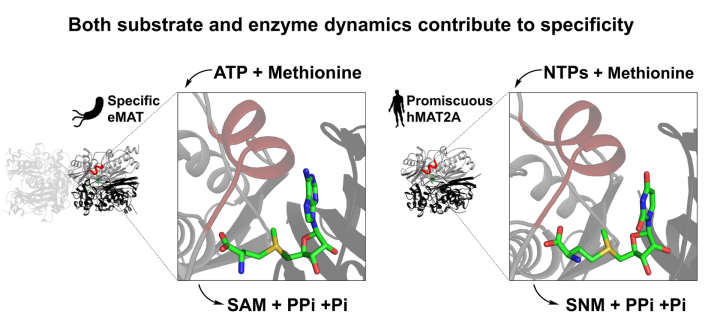
10. Gade, M. H.; Lynn Tan, L.; Damry, A. M.; Sandhu, M.; Brock, J. S.; Delanay, A.; Villar-Briones, A.; Jackson, C. J.*; Laurino P.* Substrate dynamics contributes to enzymatic specificity in human and bacterial methionine adenosyltransferases. Journal of American Chemical Society Au 2021, 1, 12, 2349-2360 https://pubs.acs.org/doi/10.1021/jacsau.1c00464
9. Clifton, B. E.; Fariz, M. A.; Uechi, G.; Laurino, P.* Evolutionary repair reveals an unexpected role of the tRNA modification m1G37 in aminoacylation. Nucleic Acid Res. 2021, 49, 21, 12467-12485 https://doi.org/10.1093/nar/gkab1067
8. Testa A.†; Dindo, M.†; Rebane, A.; Nasouri, B.; Style, R.; Golestanian, R.; Dufresne, E.*; Laurino, P.* Sustained Enzymatic activity and Flow in Crowded Protein Droplets Nature Communications 2021, 12, 6293 https://rdcu.be/cAzZP
7. Chouhan, B. P. S.; Gade, M.; Martinez, D.; Toledo-Patino S.; Laurino P.* Implications of divergence of Methionine AdenosylTransferase in archea. FebsOpenBio 2022, 12, 130-145 https://febs.onlinelibrary.wiley.com/doi/10.1002/2211-5463.13312
6. Laurino, P.* & Tokuriki N.* Dan Salah Tawfik (1955-2021). Nature Chemical Biology 2021, 17, 930. https://www.nature.com/articles/s41589-021-00864-w
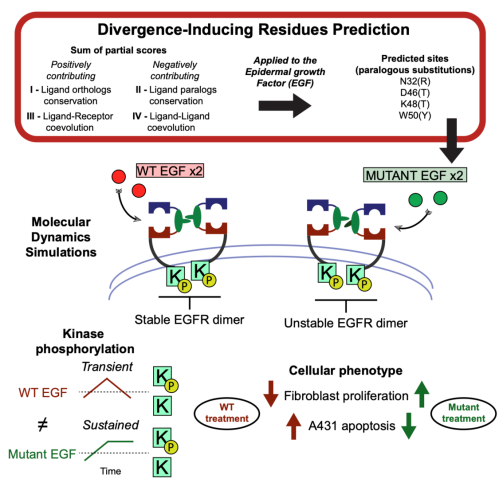
5. Pascarelli, S.; Merzhakupova, D.; Uechi, G.; Laurino P.* Single EGF mutants unravel the mechanism for stabilization of Epidermal Growth Factor Receptor (EGFR) system. Juornal of Biological Chemistry 2021, 297, 100872. https://www.jbc.org/article/S0021-9258(21)00672-4/fulltext
2020
4. Danielson, E.*; Dindo, M.; Porkovich, A. J.; Kumar, P.; Wang, Z.; Jain, P.; Mete, T.; Ziadi, Z.; Raghavendra, K.; Laurino, P.; Sowwan, M. Non-Enzymatic and Highly Sensitive Lactose Detection Utilizing Graphene Field-Effect Transistors. Biosensors and Bioelectronics. 2020, 165, 112419 https://doi.org/10.1016/j.bios.2020.112419
2019
3. Dindo, M.; Grottelli, S.; Annunziato, G.; Giardina, G.; Pieroni, M.; Pampalone, G.; Faccini, A.,;Cutruzzola, F.; Laurino, P.; Costantino, G.; Cellini, B. Cycloserine Enantiomers Are Reversible Inhibitors of Human Alanine:Glyoxylate Aminotransferase: Implications for Primary Hyperoxaluria Type 1. Biochemical Journal, 2019, 476, 3751-3768. doi:10.1042/BCJ20190507
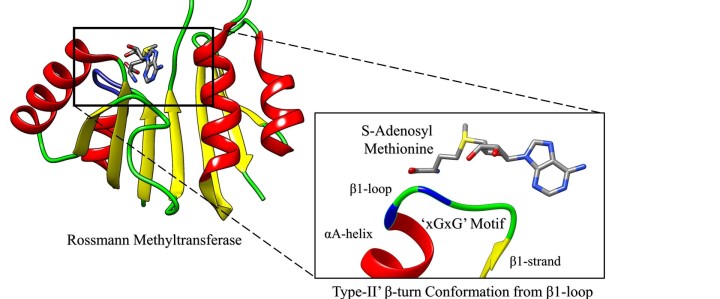
2. Chouhan, B. P. S.; Maimaiti, S.; Gade, M.; Laurino P.* Rossmann-fold Methyltransferases: taking a 'beta-turn' around their cofactor S-Adenosylmethionine. Future of Biochemistry special Issue of ACS Biochemistry, 2019, 58, 166-170. https://pubs.acs.org/doi/10.1021/acs.biochem.8b00994
2017
1. Laurino P., Tawfik D. S. Spontaneous Emergence of S-Adenosylmethionine and the Evolution of Methylation. Angewandte Chemie International Edition Engl. 2017, 56, 343-345. https://onlinelibrary.wiley.com/doi/10.1002/anie.201609615 This article has been recommended in F1000Prime as being of special significance in its field.
2016 or earlier
-
Goldenzweig, A.; Goldsmith, M.; Hill, S. E.; Gertman, O.; Laurino, P.; Ashani, Y.; Dym, O.; Unger, T.; Albeck, S.; Prilusky, J.; Lieberman, R. L.; Aharoni, A.; Silman, I.; Sussman, J. L.; Tawfik, D. S.; Fleishman S. J. "Automated computational design of human enzymes for high bacterial expression and stability". Mol Cell 2016, 63, 337-346.
-
Laurino, P.; Rockah-Shmuel, L.; Tawfik D. S. “Engineering and directed evolution of DNA methyltransferases” Adv Exp Med Biol 2016, 945, 491-509
-
Laurino, P.; Tóth-Petróczy, A.; Meana-Pañeda R.; Lin W.; Truhlar D. G.; Tawfik, D. S. “An ancient fingerprint of coevolution of the Rossmann fold and adenosine cofactors”. PLoS Biology 2016, 3, e1002396.
-
Alcolombri,U.; Laurino, P.; Lara-Astiaso, P.; Vardi, A.; Tawfik D. S. “DddD is a CoA-Transferase/Lyase Producing Dimethylsulfide in the Marine Environment” Biochemistry 2014, 34, 5473–5475. This article has been chosen to be highlighted on the journal’s home page.
-
Fumagalli, L.; Pallavicini, M.; Budresi, R.; Bolchi, C.; Canovi, M.; Chiodini, G.; Gobbi, M.; Laurino, P.; Micucci, M.; Straniero, V.; Valoti, E.; “6-Methoxy-7-benzofuranoxy and 6-Methoxy-7-indolyloxy Analogues of 2-[2-(2,6-Dimethoxyphenoxy)ethyl]aminomethyl-1,4-benzodioxane (WB4101):1 Discovery of a Potent and Selective α1D-Adrenoceptor Antagonist” J. Med. Chem. 2013, 16, 6402–6412.
-
Azzouz, N.; Kamena, F.; Laurino, P.; Kikkeri, R.; Mercier, C.; Cesbron-Delauw, M-F.; Dubremetz, J-F.; De Cola, L.; Seeberger, P. H. “Toxoplasma gondii secretory proteins bind to sulfated heparin structures.” Glycobiol., 2013, 1, 106–120.
-
Suzuki, Y; Laurino, P.; McQuade D. T.; Seeberger P. H. “A Capture and Release Catalytic Flow System”, Helv. Chim. Acta 2012, 12, 2578–2588.
-
Laurino, P.; Hernandez, H.; Bräuer, J.; Grützmacher, H.; Tauer, K.; Seeberger, P. H. “Snowballing Radical Generation Leads to Ultrahigh Molecular Weight Polymers” Macromol. Rapid Commun. 2012, 20, 1770–1774.
-
Laurino, P.; Kikkeri, R.; Seeberger, P. H. “Continuous-flow reactor–based synthesis of carbohydrate and dihydrolipoic acid–capped quantum dots” Nat. Prot. 2011, 6, 1209–1220.
-
Laurino, P.; Kikkeri, R.; Azzouz, N.; Seeberger, P. H. “Detection of Bacteria Using Glyco-Dendronized Polylysine Prepared by Continuous Flow Photofunctionalization” Nano Lett., 2011, 11, 73–78.
-
Kikkeri R.; Hong, S. Y.; Grünstein, D.; Laurino, P.; Seeberger, P. H. “Carbohydrate-based Nanoscience: Metallo-glycodendrimers and Quantum Dots as Multivalent Probes“ Beilstein Bozen Symposium "Functional Nanoscience" May 17th–21st, 2010.
-
Christina, D.; Laurino, P.; Azzouz, N.; Seeberger, P. H. “Accelerated Continuous Flow RAFT polymerization” Macromol. 2010, 43, 10311–10314.
-
Bernardes, G. J. L.; Kikkeri, R.; Maglinao, M.; Laurino, P.; Collot, M.; Hong, S. Y.; Lepenies, B.; Seeberger, P. H. “Design, synthesis and biological evaluation of carbohydrate functionalized cyclodextrins and liposomes for hepatocyte-specific targeting” Org. Biomol. Chem. 2010, 21, 4987–4996.
-
Kikkeri, R.; Laurino, P., Odedra, A.; Seeberger, P. H. “Synthesis of carbohydrate-functionalized quantum dots in microreactors”. Angew. Chem. Int. Ed. Engl. 2010, 49, 2054–2057.
-
Laurino, P.; Odedra, A.; Mak, X. Y.; Gustafsson, T.; Geyer, K.; Seeberger, P. H. “Microfluidic devices for organic processes”. Chemical Reactions and Processes under Flow condition (RSC publishing 2010).
-
Mak, X. Y.; Laurino, P.; Seeberger, P. H. “Asymmetric reactions in continuous flow”. Beilstein J. Org. Chem. 2009, 5, 1–11.
-
Mak, X. Y.; Laurino, P.; Seeberger, P. H. “Synthesis of pharmaceutical and bio-active compounds in microreactors” Chemistry Today 2009, 27, 15–18.
-
Kikkeri, R.; Lepenies, B.; Adibekian, A.; Laurino, P.; Seeberger P. H. “In vitro imaging and in vivo liver targeting with carbohydrate capped quantum dots”. J. Am. Chem. Soc. 2009, 131, 2110–2112.