Abstract
In the past few decades protein engineering allowed to generating artificial enzymes able to catalize unnatural reactions but also to become importnat tools for synthetic biology. Our Unit is particulary interested in generating new enzymes by directed evolution but also to understand how enzymes and in general proteins evolve in Nature. In FY18 we focused most of our attention on protein promiscuities, one of the starting point of enzyme evolution (in nature and in the lab).
1. Staff
- Dr. Madhuri Gade, Researcher
- Dr. Babak Bakhshinejad, Researcher
- Dr. Bhanu Chouhan, Researcher
- Dr. Mirco Dindo, Researcher (from March)
- Shayida Maimaiti, Technician
- Dalmira Merzhakupova, Graduate student
- SarahYukie Nagasawa, Graduate Student (from May to August)
- Stefano Pascarelli, Graduate Student (from September to December)
- Santanu Mondal, Graduate Student (from Septermber to December)
- Angela Kirykowicz, Graduate Student (from January to April)
- Sachie Matsuoka, Research Unit Administor
2. Collaborations
Topic: Understanding cofactor specificity for SAM analogues
- Type of collaboration: Joint research
- Researchers:
- Professor Colin Jackson, ANU, Australia
Topic: Protein Evolution after genome duplication: case study of EGFR in fish lineage
- Type of collaboration: Joint research
- Researchers:
- Professor Federica Di Palma, Earlham Institute, UK
Topic: Are epistatic patches in Rossmann Fold enzymes conserved?
- Type of collaboration: Joint research
- Researchers:
- Professor Michael Laessig, Cologne University, Germany
Topic: Importance of the TFBS surrounding in the TF search
- Type of collaboration: Joint research
- Researchers:
- Professor Simone Pigolotti, OIST
- Dr. Massimo Cencini, CNR (Rome, Italy)
3. Activities and Findings
3.1 Understanding enzymes specificities for SAM analogues
Our focus is on the enzymes which bind S-Adenosyl methionine (SAM). S-Adenosyl methionine (SAM) is very abundant and important cofactor in the cell. SAM is synthesised intracellularly by SAM Synthetase starting from ATP and methionine. Methyltransferases enzyme use SAM as methyl donor to methylate different substrates such as DNA, RNA, small molecules and lipids. Our aim is to understand the specificity of SAM binding enzymes for SAM analogues, in particular we are interested in SAM synthetase and methyltransferase enzyme. We initially studied SAM synthetases from the three kingdoms and found that SAM synthases from eukarya and archaea are able to accept different SAM analogues despite their low homology in sequence. Our findings highlight that despite enzymes might have high sequence and structure homology this conservation does not ensure same substrate specificity. Currently we are investigating the promiscuoties of SAM methyltransferases.
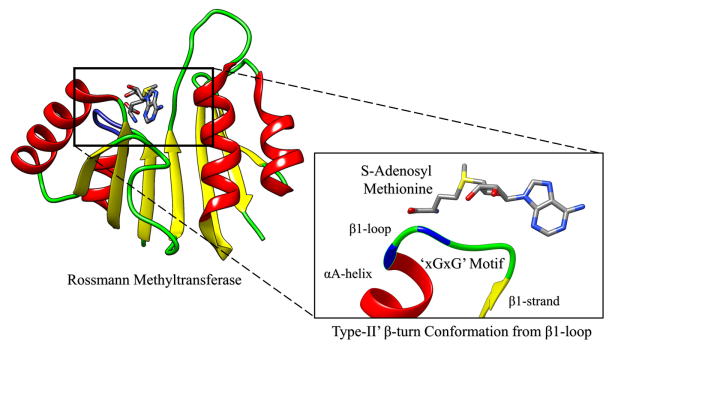
3.2 Identification of an Highly Conserved Loop in Rossmann Fold Methyltransferases
[Biochemistry, 58, 166-170, (2019)]
Methyltransferases (MTases) are superfamilies of enzymes that catalyze the transfer of a methyl group from S-adenosyl Methionine (SAM) - a nucleoside-based cofactor to a wide variety of substrates such as DNA, RNA, protein, small molecules and lipids. Depending upon their structural features, the MTases can be further classified into different classes – wherein, we consider exclusively the largest class of MTases which are the Rossmann-fold MTases. It has been shown that the nucleoside-cofactor binding Rossmann enzymes particularly the nicotinamide adenine dinucleotide (NAD)-, flavin adenine dinucleotide (FAD)-, and SAM-binding MTases enzymes share common binding motifs which includes: a Gly-rich loop region that interacts with the cofactor, and a highly conserved acidic residue (Asp/Glu) that interacts with the ribose moiety of the cofactor. Here, we observe that the Gly-rich loop region of the Rossmann MTases adapts a specific type-II’ β-turn in close proximity to the cofactor (< 4Å) and it appears to be a key feature of these superfamilies. Additionally, we demonstrate that the conservation of this β-turn could play a critical role in the enzyme-cofactor interaction, thereby shedding new light on the structural conformation of Gly-rich loop region from Rossmann MTases.
3.3 Protein Evolution after genome duplication: case study of EGFR in fish lineage
The phylogenetic tree of all ErbB receptors in fish shows a high degree of heterogeneity, with frequent domain rearrangements. We calculated dN/dS ratio based on the codon sequence, that showed the possibility of burst after duplication hypothesis. Analysis of the expression data reveals a trend in the EGFR-associated genes expression levels. The duplicated EGFRs association splitted in two clades and this division seems to follow the phylogenetic tree of the species. Among the genes that drive the clustering, one of the ligands of EGFR - HBEGF was particularly interesting. The expression of this gene is found to correlate with EGFRa only. For this reason, HBEGF is a good candidate of sub- or neo-functionalization in the system. We are currently further studying this system.
3.4 A single mutation in EGF affects the downstream EGFR pathway
Molecular co-evolution is a key feature of biological systems. Molecular interactions (ligand-receptor, protein-protein, etc.) usually evolve simultaneously and independently to optimize binding. Frequently, these interactions involve one receptor that binds multiple ligands. Each ligand often leads to a different pathway activation intra-cellularly. Understanding single amino acid roles in evolving ligands and their contributions to downstream pathways of the receptor is still challenging.
We developed a cross-conservation approach to identify functionally important EGF residues. Four EGF mutants (N32R, D46R, K48T, W50Y) have been selected and studied biochemically and at the cellular level. While these mutants retain binding affinities for EGFR similar to that of EGF, surprisingly the effects of two of them (D46R, K48T) at the cellular level changed, inducing higher proliferation levels in normal fibroblasts and reducing proliferation in skin cancer cells. These results lay the base to understand the role of mutations in diseases and to design ligand analogous for therapeutics.
4. Publications
4.1 Journals
Chouhan, B. P. S., Maimaiti, S., Gade, M. & Laurino, P. Rossmann-Fold Methyltransferases: Taking a "beta-Turn" around Their Cofactor, S-Adenosylmethionine. Biochemistry Epub ahead of print, doi:10.1021/acs.biochem.8b00994 (2018).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
Seminars
- Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering University of Cologne, Germany (2018).
- Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering, Earlham Institute, UK (2018).
- Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering, University of Insubria, Italy (2018).
Conference Oral Presentations
- Laurino, P. & Gade, M. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering, Pavia, Italy (2018).
- Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering, Institute for Molecular Science, National Institute for Natural Sciences, Aichi (2019).
- Gade, M., Villar-Briones, A. & Laurino, P. Understanding enzymes specificities as a tool for cofactor engineering, Chongqing, China (2018).
- Gade, M. & Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering, Freiburg, Germany (2018).
- Gade, M. & Laurino, P. Understanding enzymes specificities as a tool for cofactor engineering, Heidelberg, Germany (2018).
Conference Poster Presentations
- Laurino, P. A journey from an ancient finger print of Rossmann fold enzymes to cofactor engineering Yokohama, Japan (2018).
- Chohan, B. P., Maimaiti, S., Gade, M. & Laurino, P. Rossmann-Fold Methyltransferases: Taking a ‘beta-Turn’ around Their Cofactor S-Adenosyl Methionine, University of Ryukyus, Japan (2018).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar "Dynamics and Constraints of Enzyme Evolution"
- Date: July 18, 2018
- Venue: C700, OIST Campus Lab3
- Speaker: Dr. Nobuhiko Tokuriki (University of British Columbia)
6.2 Seminar "New Roles of Maintenance Methylation Factors DNMT1 and NP95/UHRF1 for Chromatin Mediated Transcriptional Regulation"
- Date: August 02, 2018
- Venue: C756, OIST Campus Lab3
- Speaker: Dr. Jafar Sarif (RIKEN IMS)
6.3 Seminar "Structures and antifungal activities of plant chitinases"
- Date: October 26, 2018
- Venue: C740, OIST Campus Lab3
- Speaker: Dr. Toki Taira (University of the Ryukyus)
6.4 Seminar "Artificial cell reactor technology for digital bioassay, enzyme screening, and system reconstitution"
- Date: November 06, 2018
- Venue: D015, OIST Campus Lab1
- Speaker: Dr. Hiroyuki Noji (The University of Tokyo)
6.5 Seminar "How simple peptides enhance RNA polymerase ribozyme function"
- Date: December 11, 2018
- Venue: D015, OIST Campus Lab1
- Speaker: Dr. Shunsuke Tagami (RIKEN Center for Biosystems Dynamics Research)
6.6 Seminar "Design and evolution of artificial enzymes"
- Date: March 07, 2019
- Venue: C700, OIST Campus Lab3
- Speaker: Dr. Donald Hilvert (ETH Zurich)
7. Other
Ecology & Evolution Seminar Series 3 : "Towards an ancient finger print of Rossmann fold enzymes and the evolution of SAM methylation"
- Date: April 18, 2018
- Venue: C700, OIST Campus Lab3
- Speaker: Dr. Paola Laurino
Internal Seminar: "Rossmann-Fold Methyltransferases: Taking a 'Beta-Turn' around Their Cofactor, S-Adenosylmethionine"
- Date: February 01, 2019
- Venue: C700, OIST Campus Lab3
- Speaker: Dr. Bhanu Chouhan
Teaching at Skill Pill
- Terminal - May 2018 (Stefano Pascarelli)
- BLAST - July 2018 (Bhanu Chouhan, Stefano Pascarelli)
- Regular Expressions - August 2018 (Stefano Pascarelli)