Research
Design of proteins with electron transfer capabilities
Engineering protein wires to power biocatalysis and bioremediation in electroactive microbes could enable faster and more controlled biochemical reactions. These wires could allow for the targeted redirection of electron flow within bacterial systems, improving the performance of bacterial factories and supporting energy-intensive processes without disrupting core metabolism. This technology aims to advance biofuel production, bioremediation, and other sustainable applications by optimizing the catalytic efficiency and energy management of engineered microbes.
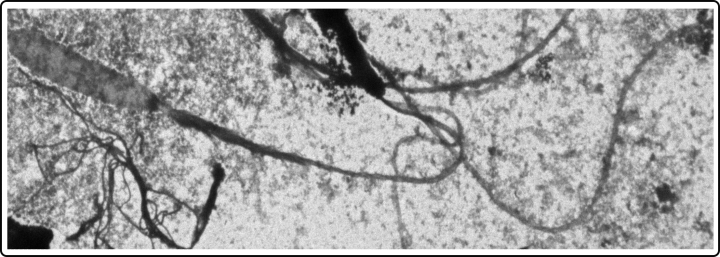
Evolution of thread-like stinging proteins in cnidaria
The evolution of protein threads, such as the stinging wires of nematocysts, represents a remarkable example of protein innovation driven by ecological pressures like predation and defense. These structures likely emerged from ancestral fibrous proteins, which were gradually adapted for specialized mechanical functions through processes like gene duplication and modular evolution. Over time, natural selection favored proteins with increased elasticity and strength, enabling the rapid, forceful discharge required to capture prey and deliver venom. Studying the molecular evolution and biomechanics of these protein threads offers valuable insights into the broader mechanisms of protein adaptation and functional diversification.
Previous reseach
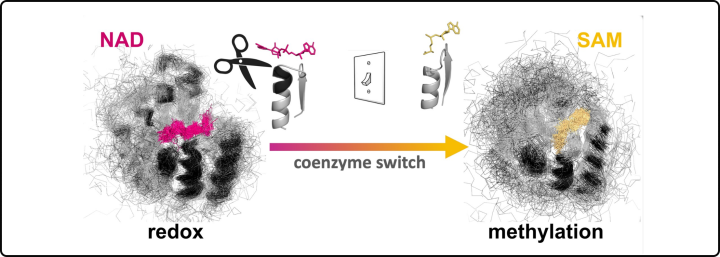
InDels mediating coenzyme-binding divergence
In this study, we focused on understanding how the most catalytically-diverse enzymes (Rossmann enzymes) switch their coenzyme preferences. Enzymes are the molecular machines that facilitate chemical reactions in our cells, and coenzymes are small molecules that help enzymes do their job. We removed three specific amino acids (the building blocks of proteins) from Rossmann enzymes to switch the binding of a redox coenzyme towards a methylating one. These findings demonstrate that enzymes likely explored distinct chemistries with minor changes in their amino acid sequences (Toledo-Patino et al, PNAS 2022).
InDels mediating structural transformations
In this study, we explored how different protein structures, evolved from common ancestors.
Proteins are made up of a limited number of building blocks, and their diverse functions and shapes come from combining these building blocks in various ways.We discovered evidence suggesting that a specific protein fold, called HemD-like, emerged from another fold called flavodoxin-like, through a process involving gene duplication and segment swapping. To test our hypothesis, we reversed these evolutionary steps, recreating the flavodoxin-like shape from a HemD-half. This experimental result strongly supported our idea of a common ancestry for these two folds (Toledo-Patino et al, Biochemistry 2019).
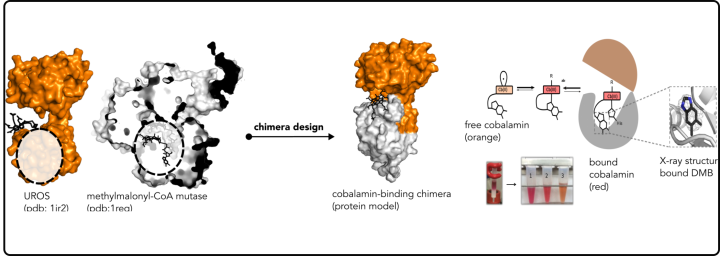
Protein-LEGO finding the puzzles of Nature
In this study we systematically searched for common fragments between proteins with distinct overall architecture, following the hypothesis that nature employed a “Lego-brick” strategy to build multiple architectures.
zInDels mediating structural transformations
We discovered fragments that are present throughout the protein universe, appearing in diverse environments. Remarkably, we identified over a thousand sub-domain sized fragments that Nature has ingeniously repurposed to create new proteins (Ferruz et al, JMB 2020). These fragments offer an exciting and innovative avenue for protein design (Toledo-Patino et al, FEBS Letters 2024 & Toledo-Patino 20219).