Coenzyme Switch: Researchers Recreate the Way Nature Changes Coenzyme Binding on Rossmann Enzymes
Nucleobase-containing coenzymes are believed to be the relics of an ancient RNA world and can provide information on the origin and evolution of proteins. However, coenzyme-protein interactions largely remain unclear. Recently, researchers from the Okinawa Institute of Science and Technology Graduate University looked at Rossmann enzymes for answers, discovering that insertions and deletions essentially mold coenzyme specificity in these proteins. Their findings, while evolutionarily significant, also potentially provide a novel strategy to engineer coenzyme specificity.
Coenzymes are molecules that assist proteins in nearly half of all the reactions they catalyze. These small, organic molecules contain nucleotides just like in the building blocks of our DNA and RNA. While coenzymes play an extremely crucial role in the catalysis of proteins, their importance is not limited to this alone. Nucleobase-containing coenzymes are considered the fossil remnants of an ancient RNA-based world, which has been hypothesized to exist even before the very first proteins came into existence. They can, hypothetically, offer a closer look at how proteins emerged and evolved. Unfortunately, not much is known about the evolution of coenzyme-protein interactions.
The structure and function of any protein is coded in its amino acid sequence. Certain structures are evolutionarily conserved across all kingdoms of life. One such recurring structure—the “Rossmann fold”—was discovered by Dr. Michael Rossmann in 1970. The Rossmann fold is the most catalytically diverse and abundant protein architecture in nature and is an excellent target to study coenzyme-protein interactions. Rossmann proteins display diverse catalytic functions owing to minuscule differences in their structure. These differences, affect the binding specificities of their co-acting catalysts, coenzymes.
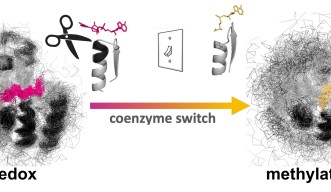
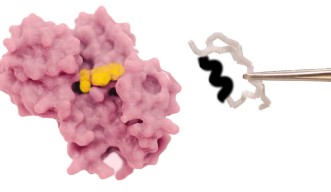
Researchers from Okinawa Institute of Science and Technology (OIST) recently modified the coenzyme pocket of a Rossmann protein that naturally performs redox reactions to bind a methylating agent instead. While the natural protein binds to the coenzyme NAD (nicotinamide adenine dinucleotide), the mutant lost the ability to bind NAD and acquired binding for SAM (S-adenosyl methionine). Their findings have been published in Proceedings of the National Academy of Sciences of the United States of America.


Says Dr. Paola Laurino, Assistant Professor who leads OIST Protein Engineering and Evolution Unit, “As proof of principle, we engineered an oxidoreductase protein to accept a methylating coenzyme “SAM” instead of its natural coenzyme “NAD.” This involves the redesign of the ancient and highly conserved glycine rich loop. This task is not trivial because of the complexity of the intramolecular H-bonding and it has attracted the attention of many protein engineers in the past.”
Historically, protein interactions have been studied using site-directed mutagenesis—a process that gives rise to modified proteins where one or more amino acids are substituted with others. However, until now, researchers have not fully unleashed the potential of “insertions” and “deletions” (collectively referred to as “InDels”). In contrast to amino acid substitutions, an “InDel” significantly modifies protein structure because of the addition or removal of one or more amino acids from the corresponding protein sequence.
First, the researchers performed extensive analyses and reshaped the ancient coenzyme-binding motif of NAD into the SAM-binding one. To achieve this, they removed an InDel of three amino acids from the NAD coenzyme pocket and solved the structure of the resulting mutant. As expected, the mutant exhibited the characteristic structural features of a SAM-binding pocket.
Next, the team decided to validate their finding by studying the interactions of the generated mutant with SAM. To this end, the team performed isothermal titration calorimetry measurements—a biophysical technique that determines binding affinities—and validated the successful coenzyme switch when they observed that the mutant was indeed binding. The results were further corroborated through computerized simulations.
Lead author Dr. Saacnicteh Toledo-Patiño, a postdoctoral researcher at OIST Protein Engineering and Evolution Unit concludes, “It is amazing that the sequence combinations possible for a small protein, about 100 amino acids in length, exceeds the number of atoms in the known universe. For this reason, nature has only explored an infinitesimal fragment of these possibilities, and yet, is able to drive the vast number of reactions that sustain life.”
This knowledge can aid the design of artificial enzymes with immense applications.
Research paper
- Title: Insertions and deletions mediated functional divergence of Rossmann fold enzymes
- Authors: Saacnicteh Toledo-Patiño, Stefano Pascarelli, Gen-ichiro Uechi and Paola Laurino
- Affiliations: Okinawa Institute of Science and Technology
- Journal: Proceedings of the National Academy of Sciences of the United States of America
- DOI:10.1073/pnas.2207965119
- Date: 2022.11.23
Article Information
Specialties
Research Unit
For press enquiries:
Press Inquiry Form