OIST Scientists Decode the Pearl Oyster Genome

The Marine Genomics Unit of the Okinawa Institute of Science and Technology Graduate University (OIST) in collaboration with Japanese research groups including from the University of Tokyo and Mikimoto Co., Ltd. has completed the first decoding of the genome of the Japanese pearl oyster.
Their success in deciphering the genome means that researchers now have all of the genetic information needed to understand the mechanisms of pearl and shell formation. Great progress on pearl oyster research is promised with this new genome data that will help uncover the secrets of how oysters generate beautiful pearls and shells. In addition, by comparing the black-lipped and the white-lipped pearl oysters that are cultured in Okinawa Prefecture, the researchers will learn how these oysters generate their distinctive black and gold pearls. This research provides a very useful tool to the aquaculture industry for quality control of organisms, development of high-grade pearls, and parentage analysis.
This genomic information will benefit not only research into pearl formation but also the study of pearl oyster ecology and physiology. Scientists can now analyze the impacts of environmental changes, such as increases in seawater temperature and acidification, on bivalve organisms. This information will also be very valuable in studying the influence of environmental change on other bivalve organisms, such as other species of oysters, scallops, and clams.
The Japanese pearl oyster belongs to the phylum Mollusca, which includes around 85,000 species and includes oysters, abalones, squids, cuttle fish, octopuses, and snails. The mollusks are a highly diverse group, and their habitat extends from seawater to fresh water to land. Despite the fact that many mollusks are important organisms for the fishery industry, there has previously been no comprehensive genome data of this group of animals. The collaboration’s publication is the world’s first article on the molluscan genome. The deciphered genome of the pearl oyster will provide crucial information for understanding overall molluscan biology.
Dr. Takeshi Takeuchi of the Marine Genomics Unit said, “The cultured pearl business using pearl oysters was born and nurtured in Japan. The country has also taken the lead in the investigation on the underlying mechanism behind the formation of beautiful pearls within the oyster. However, research on mollusks, including the pearl oyster, had been difficult because their genome and DNA was not readily available. This research paves the way for advancement of not only pearl research, but also research on mollusks as a whole.”
This study was published on February 7, 2012 in the online version of the scientific journal DNA Research (Oxford University Press).
Background
Genome of the Pearl Oyster
The Japanese pearl oyster, Pinctada fucata, is a mollusk of the Bivalvia class (two shells per individual). The genome size of bivalves is comparatively large, often more than 1.5 gigabases. In contrast, the size of the genome of most other animals in general ranges from 0.4 to 0.6 gigabases. This makes bivalve genome analysis difficult to accomplish, and so far no decoded genomes have been reported for mollusks. The genome of P. fucata is estimated to be 1.15 gigabases, relatively small among the bivalves but still a huge challenge for sequencing. We have successfully decoded the P. fucata genome by using so-called ‘next generation sequencers’. We found the genome contains approximately 23,000 protein-coding genes, 70% of which are supported by corresponding “expressed sequence tags,” which means that they can be detected as actively expressed genes. The genes sequenced include those reported to be associated with bio-mineralization. Genes encoding transcription factors and signal transduction molecules are present in numbers comparable with genomes of other metazoans. In addition, we found that repeat sequences, such as transposons and microsatellites, occupied 10% of the genome. Genome-wide molecular phylogeny suggests that the lophotrochozoans (a major subgroup of bilaterally-symmetrical animals) represent a distinct clade from the ecdysozoans (including animals such as the arthropods). The present decoding of the pearl oyster genome thus provides a platform for the identification of selection markers and genes for calcification, knowledge of which will be important in the pearl industry.
Revealing the mechanism of pearl formation
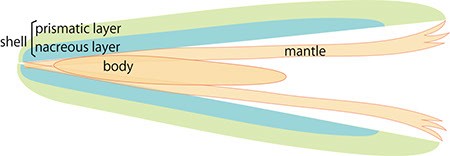
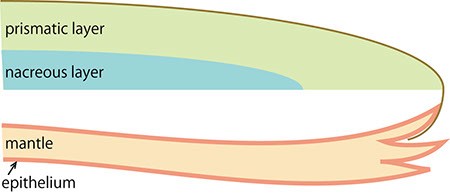
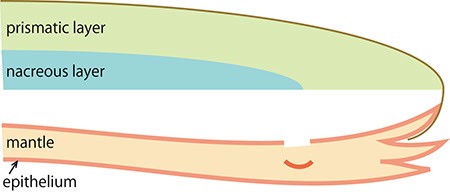
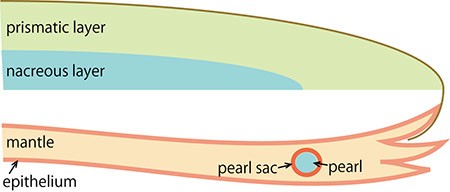
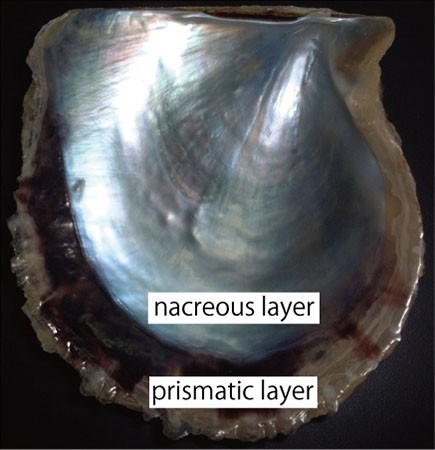
The pearl oyster has an organ called the mantle that secretes proteinous organic materials and forms shells (Figure 1A and B). The luster layer in the inner part of the shell is called the ‘nacreous layer’ (Figure 2). When the surface of the mantle is injured and some of its cells fall into the inner part of the tissue (Figure 1C), the cells sometimes proliferate to form a sac-like structure. These cells may then secrete proteins and a hard material, which is made of identical components to the shell, thus forming a pearl (Figure 1D). The nacreous layer is comprised of fine alternating layers of calcium carbonate about 0.4 micrometers thick and a thin organic layer about 0.02 micrometers thick (Figure 3). It is not yet understood how pearl oysters form such a fine structure. In addition, the nacreous layer is 1000 times tougher than pure calcium carbonate crystal, which has caught the attention of material scientists.
The other structure in the pearl oyster shell is the ‘prismatic layer’ (Figure 2). Pearl oysters are thought to secrete specific proteins selectively to form these two different shell layers, but the precise mechanism of generating these distinct structures is not known.
As mentioned above, proteins play important roles in both pearl and shell formation, and we found in the genome several genes reported to have an association with bio-mineralization. In other words, our success in deciphering the pearl oyster genome means that we have obtained all of the genetic information for understanding the mechanisms of pearl/shell formation. Much progress in pearl oyster research is promised as a result of the genome data, and we may uncover the secrets of how they generate the beautiful pearls and shells. In addition, comparing the black-lipped and the white-lipped pearl oysters that are cultured in Okinawa Prefecture will show us how these oysters generate beautiful pearls with various colors such as black and gold.
Fundamental information for pearl oyster quality control
Our team found specific repetitive DNA sequences such as transposons and microsatellites, which occupy about 10% of the pearl oyster genome. These sequences are available as DNA markers, providing very useful tools in aquaculture industry for quality control of organisms, development of high grade pearls, and parentage analysis.
The Pearl in Japan
The Pearl appeared in the 8th Century Man-Yo-Shu, Japan’s oldest anthology of Tanka poetry, indicating that the jewel has been beloved of the Japanese since ancient times. In 1893, Kokichi Mikimoto was the first in the world to succeed in aquaculture pearl production using the Japanese pearl oyster, Pinctada fucata. Since then, pearl culture has become one of the most important aquaculture industries in Japan.
However, the Japanese pearl culture industry suffered serious damage from the occurrence of red tides in the early 1990s and red coloration disease in the late ’90s. Furthermore, rapid cross-breeding between Japanese and southern pearl oysters has taken place in recent years, resulting in reduced production of high-quality pearls, which are only produced by domestic pearl oysters.
Therefore, the researchers used a domestic pearl oyster strain that is preserved by the Pearl Research Institute, Mikimoto Co., Ltd, for the pearl oyster genome-sequencing project. The genome will provide the fundamental information for understanding the secret of brilliant Japanese pearls and for the conservation of the domestic strain.
The luster of the pearl has always attracted people; equally the natural phenomenon of pearl formation is fascinating for the scientist. Biomineralization is the process by which an organism generates hard parts, which in pearl oysters form both pearls and shells. Japan is the world leader in pearl oyster biomineralization research. Most of the proteins involved in shell and pearl formation were discovered by Japanese scientists (Miyamoto et al., 1996, Tsukamoto et al., 2004, Suzuki et al., 2009, Kinoshita et al., 2011). Considering this historical and scientific background, it is highly significant that it was a Japanese research team that finally decoded the domestic pearl oyster genome. This genome information will contribute to biomineralization research, and the Japanese pearl oyster is expected to become the “model organism of biomineralization”.
Research details
The domestic pearl oyster cultured at the Pearl Research Institute of Mikimoto Co., Ltd was used. Sperm used for the analysis was obtained from a single male. The genome size was estimated to be approximately 1.15 Gbp, one third of the human genome, by flow cytometry measurement. The researchers used two different high throughput DNA sequencers at OIST, a Roche 454 GS-FLX and an Illumina GAIIx, and the resulting data reached more than 40 times full coverage of the pearl oyster genome. The genomic sequence was reconstructed from the overlapping fragment sequences by high-performance cluster computer to obtain the pearl oyster genome assembly. The estimated number of genes in the genome is 23,257 using gene prediction software. The genome and gene data are available on the OIST website.
It is presumed that the genome data include genes involved in pearl and/or shell formation. OIST scientists confirmed that the genome contains all of the pearl and or shell forming genes that have been previously reported from P. fucata. On the other hand, the shell forming genes found in abalone, a fairly distant relative to the bivalve oyster lineage, were not present in the pearl oyster genome. This indicates the pearl oysters and the abalone use different molecular mechanisms for the formation of their nacreous layer.
The OIST group also found specific repetitive DNA sequences such as transposons and microsatellites, which occupy about 10% of the pearl oyster genome. These sequences have good potential to be used as DNA markers and will be very useful tools in the aquaculture industry for quality control of organisms, development of high grade pearls, and parentage analysis.
OIST researchers organized the “Pearl Oyster Genome Jamboree” in May 31 ~ June 2, 2011 at OIST and on January 17 ~ 19, 2012 at Tokyo University. In the Genome Jamboree, researchers of the pearl oyster and the other mollusks analyzed the genome data, shared results, and discussed their findings. In both meetings, many researchers from around Japan were involved, with around 200 participants. Through these and other activities using this newly-published pearl oyster genome, the study of pearl oyster and mollusks will be greatly advanced.
Journal and the date of publication:
DNA Research February 7, 2012 (online version)
Title:
Draft Genome of the Pearl Oyster Pinctada fucata: A Platform for Understanding Bivalve Biology
Authors:
Takeshi Takeuchi1, Takeshi Kawashima1, Ryo Koyanagi1, Fuki Gyoja1, Makiko Tanaka1, Tetsuro Ikuta1, Eiichi Shoguchi1, Mayuki Fujiwara1, Chuya Shinzato1, Kanako Hisata1, Manabu Fujie1, Takeshi Usami1, Kiyohito Nagai2, Kaoru Maeyama3, Kikuhiko Okamoto3, Hideo Aoki4, Takashi Ishikawa5, Tetsuji Masaoka6, Atushi Fujiwara7, Kazuyoshi Endo8, Hirotoshi Endo9, Hiromichi Nagasawa9, Shigeharu Kinoshita9, Shuichi Asakawa9, Shugo Watabe9, and Nori Satoh1
- Okinawa Institute of Science and Technology Graduate University
- Pearl Research Institute, Mikimoto Co., Ltd
- Mikimoto Pharmaceutical Co., Ltd
- Mie Prefecture Fisheries Research Institute
- Faculty of Bioresources, Mie University
- Aquaculture Technology Division, National Research Institute of Aquaculture, Fisheries Research Institute
- Research Center for Aquatic Genomics, National Research Institute of Fisheries Science
- Graduate School of Science, University of Tokyo
- Graduate School of Agricultural and Life Sciences, University of Tokyo
Contact information
About this research
Okinawa Institute of Science and Technology Graduate University (OIST) (http://www.oist.jp)
Marine Genomics Unit
Professor: Nori Satoh
TEL: 098-966-8634 FAX: 098-921-3844 E-Mail: norisky@oist.jp
Researcher: Takeshi Takeuchi
TEL: 098-966-8653 E-Mail: t.takeuchi@oist.jp
About OIST
Okinawa Institute of Science and Technology Graduate University (OIST) (http://www.oist.jp)
Kaoru Natori
Media Section Leader, Communication and PR Division
TEL: 098-966-2389 FAX: 098-966-2887
E-Mail: kaoru.natori@oist.jp
OIST information
The Okinawa Institute of Science and Technology Graduate University (OIST) is a new graduate school established in November, 2011, aiming to conduct internationally outstanding education and research in science and technology, and thus contribute to the sustainable development of Okinawa, and promote and sustain the advancement of science and technology in Japan and throughout the world. The OIST graduate education and research program is cross-disciplinary and aims to be at the leading edge of research in science and technology, including the Life Sciences, the Physical Sciences, and Mathematics. To lay the foundation for the Graduate University, 45 Research Units (over 200 researchers), including the Marine Genomics Unit, have been launched so far, with research in five major areas of Neuroscience, Molecular, Cell, Developmental Biology, Mathematical and Computational Sciences, Environmental and Ecological Sciences, as well as Physics and Chemistry.
Figures
Figure 1.
A. Schematic cross section of pearl oyster. The body is covered by the mantle organ and a pair of shells enclose the whole body.
B. Enlarged figure of the boxed part of A.
C. Cells in the surface of the mantle fall into the tissue following an injury.
D. The migrating cells proliferate and form a sac structure (the pearl sac). The cells then secrete proteins into the sac to generate the pearl.
Figure 2. Inner surface of the pearl oyster shell.
Figure 3. Scanning Electron Microscope image of the cross section of the nacreous layer.

Research Unit
For press enquiries:
Press Inquiry Form