Probing RNA Function with 10,000 Mutants
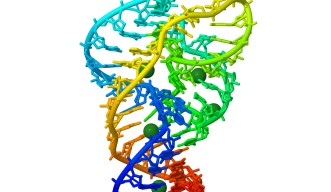
Ribozymes – ribonucleic acid enzymes – are RNA molecules that catalyse chemical reactions. Much like DNA, RNA is a linear molecule made by connecting four chemical building blocks called bases represented by the letters A, C, G, and U following the genetic information encoded in DNA.
Ribozymes are involved in key genetic processes and biochemical reactions, for example in the replication mechanism of some viruses. Scientists discovered ribozymes more than 30 years ago, but only recently these RNA structures have been detected in a wide variety of organisms, including vertebrates. These discoveries have sparked new interest among researchers, as the roles and functions of these RNA molecules are still mostly unclear.
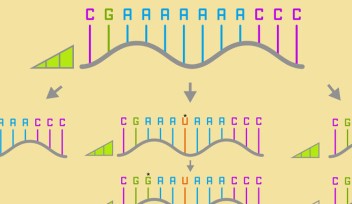
Scientists typically try to gain a better understanding of the structure and properties of a ribozyme by introducing mutations in their RNA sequence. Starting from a working ribozyme, researchers modify – or ‘mutate’ – specific bases in the ribozyme and verify if the mutated ribozyme is still capable of catalysis. However, the currently available techniques allow only a small number of mutants – several dozens – to be studied at a time. Because even a small ribozyme can have many possible mutants, researchers must select arbitrary parts of the ribozyme to make mutations, thus potentially overlooking other parts of the ribozyme that may be important for its function. Shungo Kobori and Yohei Yokobayashi, researchers in Nucleic Acid Chemistry and Engineering Unit at the Okinawa Institute of Science and Technology Graduate University (OIST), have now developed an efficient and unbiased approach to study ribozyme mutants that overcomes these limitations. Their results are published in Angewandte Chemie.
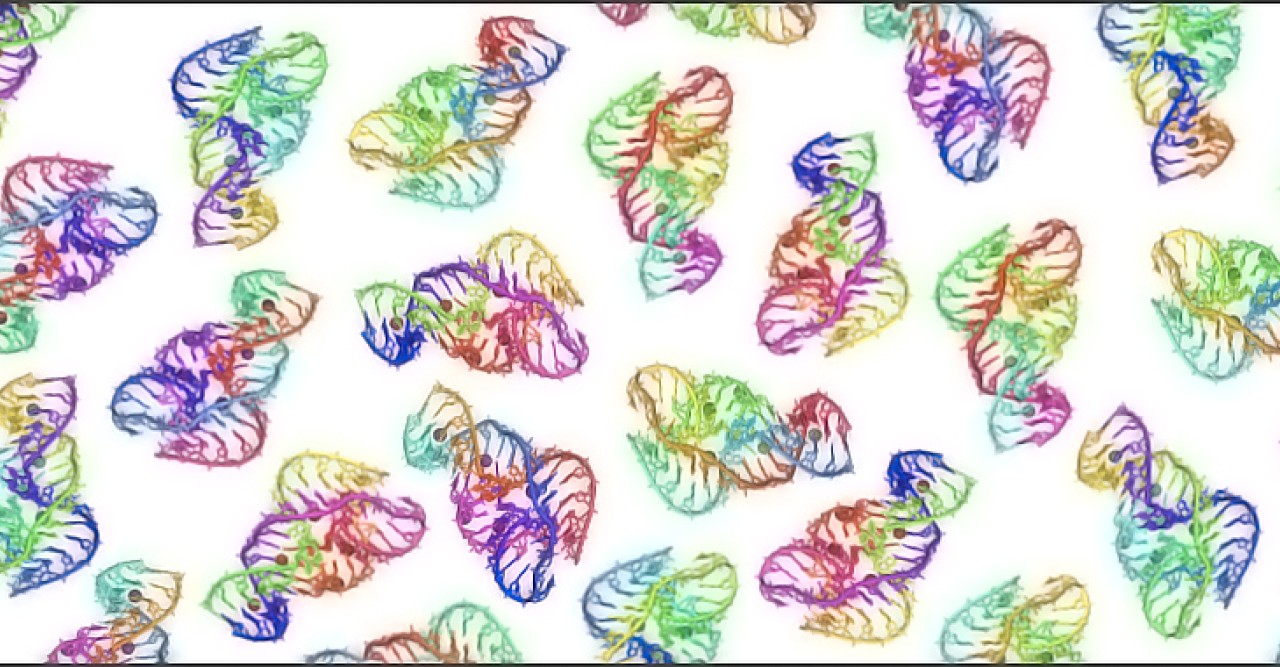
“Instead of selecting specific mutations,” Yokobayashi explained, “we decided to make and test as many mutants as possible of a specific ribozyme.” A ‘single’ mutant is a ribozyme that differs in only one base from the original ribozyme. A ‘double’ mutant differs in two bases from the original ribozyme. Even for the small ribozyme made of 48 bases studied by the researchers, there are 10,296 single and double mutants in total.
“With the help of a powerful DNA sequencer at OIST, we generated and then measured the chemical activity of all the single and double mutants of a variant of the ‘twister ribozyme’, which is found in the genome of rice” Yokobayashi said. “Thanks to this comprehensive approach, we now have a better understanding of which bases are more important for this ribozyme activity.”
A key finding is that the ribozyme is highly robust against mutations. “This result is surprising,” Yokobayashi commented, “because the ribozyme we studied has a quite compact and complex structure.” Although many of the bases are seemingly involved in keeping the intricate ribozyme structure, a significant fraction of the mutants retained detectable ribozyme activity. This capacity to tolerate mutations could have been an advantage from an evolutionary point of view, and possibly explain the widespread presence of ribozymes across different forms of life.
Deeper understanding of ribozymes could also have significant practical implications. Ribozymes can be engineered to control gene expression in living cells and viruses, with potential applications in gene therapy and regenerative medicine.
Research Unit
For press enquiries:
Press Inquiry Form