mgu FY2019 Annual Report 5.1 fig2
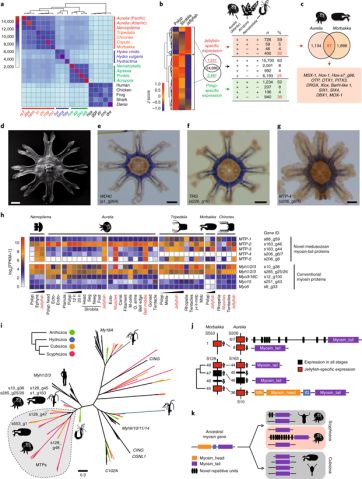
Fig. 2 | Phylogenetic distribution of genes and tissue-specific gene expression. a, Numbers of shared orthologous genes among cnidarian groups (Supplementary Table 6). Based on gene sets, there are four clusters corresponding to Anthozoa, Hydrozoa, Cubozoa and Scyphozoa. Representative bilaterians were used as an outgroup. Abbreviated genus/species names are shown at the bottom. The colours and order correspond to the genus/ species names shown on the right-hand side. The colour key shows the number of shared orthologous genes among cnidarian groups. b, Left, expression of polyp- and jellyfish-specific genes in Aurelia. Middle, a total of 1,231 genes are jellyfish-specific, 2,487 are polyp-specific and 24,886 genes do not show stage-specific expression. Right, presence or absence of Aurelia genes with stage-specific expression in the genomes of Acropora and Nematostella. BLASTP cut-off: 1 × 10−4. c, Venn diagram showing the number orthologous genes with jellyfish-specific expression in Aurelia and Morbakka. A list of 13 homeobox genes that are common markers of a jellyfish stage in Scyphozoa and Cubozoa is shown below. d, Scanning electron microscope (SEM) image of a juvenile medusa. e-g, Genes exclusively expressed in striated muscles of Aurelia revealed by in situ hybridization. WD40-repeat protein gene (s1_g264; that is, gene 264 in scaffold 1) (e), putative taxonomically restricted gene with novel repetitive domains (s226_g16) (f) and novel myosin tail protein 4 gene, MTP4 (s206_g6/7) (g). Scale bars: 200 μm. h, Expression of novel and conventional myosins in various stages and tissues of Nemopilema, Aurelia, Tripedalia, Morbakka and Chironex. s88_g59 and so on represent gene IDs in the Aurelia genome (see Supplementary Note 3.1). Numbers following ‘Myh’ or ‘Myo’ represent the classification of medusozoan myosins based on phylogenetic reconstruction and their domain composition. 12 h and 20 h represent the time after metamorphosis induction. ecto-, ectoderm; endo-, endoderm; FPKM, fragments per kilobase of transcript per million mapped reads; j-r-t-mnb, jellyfish without rhopalia, tentacles and manubrium; m-cells, mesoglea cells; meta-, metamorphosis; mnb, manubrium; muscles, ectodermal striated muscles; nseg, non-segmented part; o. arms, oral arms of a jellyfish; seg, segments. Captions for the jellyfish stages and muscle tissues are shown in red text. i, Maximum-likelihood phylogenetic tree of myosin proteins based on the alignment of their Myosin_tail_1 domains. Proteins of Aurelia and Morbakka belong to a distinct clade highlighted in grey. LG substitution model was used. Bootstrap support for all nodes is shown in Supplementary Fig. 14. j, Genomic localization of novel MTPs in the genomes of Morbakka and Aurelia. Scaffolds (S) and the genes within the scaffolds are numbered. A conventional myosin gene is located in Morbakka scaffold 128, adjacent to novel MTP genes. IQ, calmodulin-binding domain; MN, myosin N-terminal domain. k, A possible evolutionary scenario of MTP development from ancestral conventional myosin by gene duplication, followed by subsequent gene family expansions. MTPs of Aurelia and Morbakka seem to be of common origin, but have already acquired structural differences, such as additional repetitive domains in Aurelia.
Copyright OIST (Okinawa Institute of Science and Technology Graduate University, 沖縄科学技術大学院大学). Creative Commons Attribution 4.0 International License (CC BY 4.0).